Annual NC School for the Deaf Outreach Hosted on Zoom
The BIT Program’s biannual outreach event to the middle and high school science students at the NC School for the Deaf (NCSD) is always one of the highlights of the year. In early Fall, the lead science teacher, Daphne Peacock, reached out about a potential visit. Most years, the BIT instructors would make the trek to Morganton (about a three-hour drive from NC State) so that they could work with the students in person. Because of the ongoing COVID-19 pandemic, they had to Zoom in, but they sent plenty of hands-on materials for students to use during the presentation. Working with NCSD’s curriculum, the team decided to focus on two important protein functions of red and white blood cells: Teams Hemoglobin and Antibody were born!
The morning of the event, the BIT team, interpreters, and students logged into the Zoom room and talked or signed to each other as they worked out the logistics. Once all of the students had arrived for their morning science class, the presentation began as the members of NC State’s BIT Program discussed their chosen proteins and how they function within the human body. The morning started with a presentation from Dr. Stefanie Chen that discussed the basics of protein structure and function. Earlier in the week, the class had made rock candy, which Dr. Chen referred to as an example of the crystallization process needed to determine what proteins look like in 3D space, using a technique known as X-ray crystallography.
As she presented, the two interpreters took turns signing for the watching students. The live transcript function on Zoom was also enabled, though its accuracy was spotty for the more technical terms. To make sure that it was easy to focus on what was being said, the meeting was set up to spotlight only the speaker and the current interpreter.
After the initial introductory presentation, the first of the two teams took over. Team Hemoglobin and Team Antibody were each composed of a mix of postdocs and graduate students. The latter are participants in the MBTP Program, which provides funding and training for graduate students in various biotechnology-related fields, and participated as part of their outreach commitment.
Dr. Sengupta, a senior postdoc, started Team Hemoglobin’s presentation by talking about the physical structure of the hemoglobin protein. The students had been sent kits of 3D printed structures that would allow them to construct their own version of the molecule and examine the ways in which the subunits fit together. He also used Chimera, a web-based software that helps visualize 3D molecules, to show students what the molecule looked like, which helped guide them as they put together their own version of it.
Xin Dong, an MBTP graduate student who studies protein structure-function relationships in Dr. Carol Hall’s lab, took over to lead the students through the team’s worksheet activity, which contained instructions for manipulating the models to help answer the questions. They also presented slides paired with the activity to go through the biological function of hemoglobin.
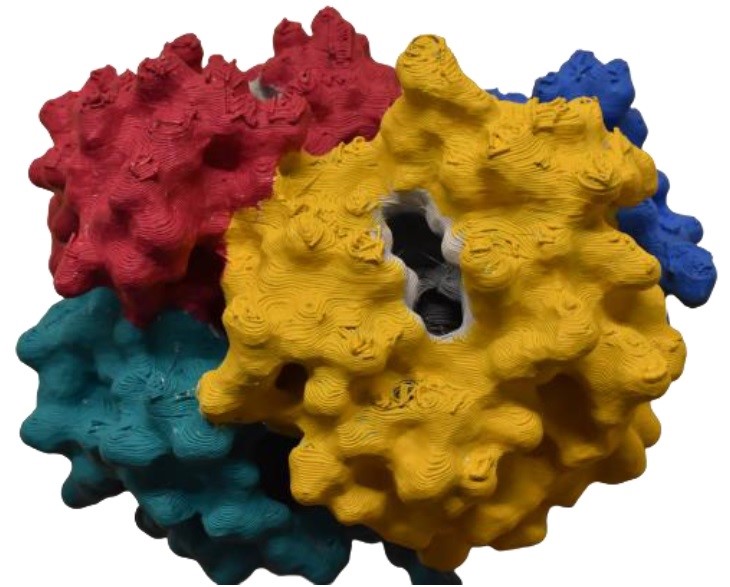
“How many oxygen molecules need to bind to the protein for it to be fully saturated?” they asked. The students used the chat to answer them, after taking a minute to sign amongst themselves. After allowing them plenty of time to conjecture, Xin revealed that the correct answer was four, and explained why.
Once the worksheet was completed, it was Team Antibody’s turn. Michael Bergman, another graduate student from the Hall lab, kicked his presentation off with an explanation of what antibodies are and what they look like. Before the event, students had already labeled the parts of the antibody, so Michael led them through additional parts of the activity, which focused on epitopes. All of the activities are available on the website that Team Antibody constructed.

As Michael explained to the students, the number of epitopes is equal to the number of different spikes present on the antibody. He encouraged students to try to figure out how many epitopes all of the example antibodies had. The science teacher spotlighted his screen for a few minutes so one of his students could sign his answer to the interpreter, instead of using the Zoom chat. Michael also played a video for them which was designed to help them visualize the 3D structure of the antibodies. The accompanying sound for the video was turned off, and the interpreters helped Michael explain anything that students saw on screen that may be confusing.
Dr. Hasley, a BIT postdoc, helped lead the students through the next part of the team’s hands-on activity to mix-and-match the epitopes on their 3D antibody models. The arms of the antibodies were connected by magnets, allowing interchangeability. By combining two shapes of the antibody “arms,” they could then recognize different antigen shapes. Students worked in groups to see how many other magnets they could get their “antibodies” to bind to.
After both teams had finished, the students were free to ask questions about all of the information that had been presented that day. While some instructors answered questions verbally through the interpreters, others addressed questions as they came up in the chat. Many of the students were curious about the antibodies produced by the COVID-19 vaccines.
One student asked (by signing through their instructor’s computer camera): Why do some people still get COVID after vaccination? Dr. Chen and Dr. Hasley worked together to explain that vaccines don’t necessarily prevent infection, and used the previous demonstration to talk about how the specificity of antibodies affects whether infection occurs or not.
Eventually, time ran out and the students had to go to their next class, but the questions continued until the very end of the event. While sending the models to the school ahead of time helped to make the event interactive, the BIT instructors hope to be able to return to NCSD in person next year.
- Categories: