Dr. Carlos C. Goller
I am an Associate Teaching Professor in the Department of Biological Sciences and teach in the Biotechnology (BIT) Program at North Carolina State University in Raleigh, NC. My research interests include molecular microbiology, metagenomics, epidemiology, history of disease, science education, and open educational practices. I am also interested in teaching with technology and the scholarship of teaching and learning.

Research Areas:
- Microbiology
- High-throughput discovery
- Molecular epidemiology
- Metagenomics and microbial genome analyses
- Metabolic modeling
- Open educational practices
Techniques:
- Bacterial culture
- DNA isolation
- qPCR & digital PCR
- High-throughput sequencing
- Bioinformatics
- Molecular cloning
- Automation & high-throughput discovery
- Metabolic modeling
Project Descriptions:
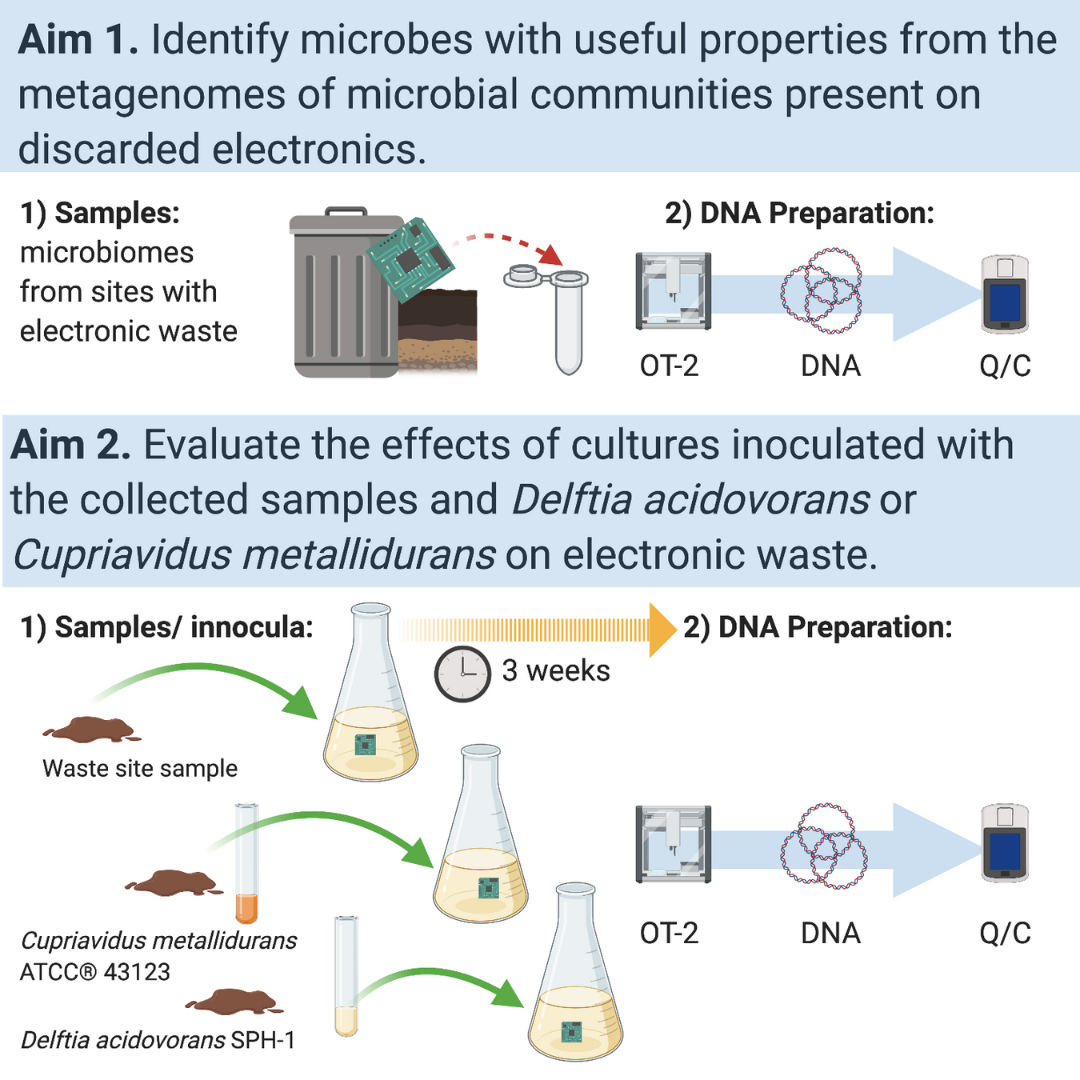
Gold and precious metals are in high demand. The increased desire for electronic components requiring gold and other metals and the environmental impact of mining present a challenge to developing sustainable sourcing methods. Fortunately, microbial communities have found ways to cope with gold toxicity by using unique genetic mechanisms to reduce toxicity and increase competitive advantage in bustling cities of microbes. One such fascinating microbe is Delftia acidovorans.
Delftia have over 50 kilobases of sequence dedicated to producing a non-ribosomal peptide that allows precipitation of gold and kills pathogens, including methicillin-resistant Staphylococcus aureus (Johnston et al. 2013, Tejman-Yarden et al. 2019). For the last five years, we have leveraged the power of citizen scientists on our campus and high-throughput (HT) approaches to catalyze the discovery and better understanding of Delftia in numerous courses at NC State and NC Central University (Riley et al. 2020).
References:
Dow EG, Wood-Charlson EM, Biller SJ, Paustian T, Schirmer A, Sheik CS, Whitham JM, Krebs R, Goller CC, Allen B, Crockett Z, and Arkin AP (2021) Bioinformatic Teaching Resources – For Educators, by Educators – Using KBase, a Free, User-Friendly, Open Source Platform. Front. Educ. 6:711535. doi: 10.3389/feduc.2021.711535
Johnston CW, Wyatt MA, Li X, Ibrahim A, Shuster J, Southam G, Magarvey NA. (2013) Gold biomineralization by a metallophore from a gold-associated microbe. Nat Chem Biol. 2013 Apr;9(4):241-3. doi: 10.1038/nchembio.1179. Epub 2013 Feb 3. PMID: 23377039.
Riley NG, Goller CC, Leggett ZH, Lewis DM, Ciccone K, Dunn RR. (2020) Catalyzing rapid discovery of gold-precipitating bacterial lineages with university students. PeerJ 8:e8925 https://doi.org/10.7717/peerj.8925
Tejman-Yarden N, Robinson A, Davidov Y, Shulman A, Varvak A, Reyes F, Rahav G, and Nissan I (2019) Delftibactin- A, a Non-ribosomal Peptide With Broad Antimicrobial Activity. Front. Microbiol. 10:2377. doi: 10.3389/fmicb.2019.02377
Image Credits:
Photos by Carlos C. Goller. Diagram created by Carlos C. Goller using BioRender.com.